Speaker
Description
Early process development in biopharma traditionally relies on small-scale experimentation, e.g. microtiter plates. At this stage, most catalyst candidates (clones) are discarded before process optimisation is conducted in bioreactors at larger scales, which differ significantly in their feeding strategies and process dynamics. This disconnect limits the representability of small-scale experiments, potentially overlooking critical interdependencies between clones and process conditions (1). To address these challenges, we introduce a multi-fidelity batch Bayesian optimisation framework that integrates a computational bioprocess model and variable-scale experimentation to improve decision-making in bioprocess development.
A key component to test the framework is a scale-aware process model for Chinese hamster ovary cells, adapted from Craven et al. (2). It captures clone-specific behaviours and simulates challenges in scaling up to pilot-scale reactors in biopharma (Figure 1). By incorporating noise, feeding strategies, and kinetics, the model generates in silico data that reflect realistic experimental conditions across scales.
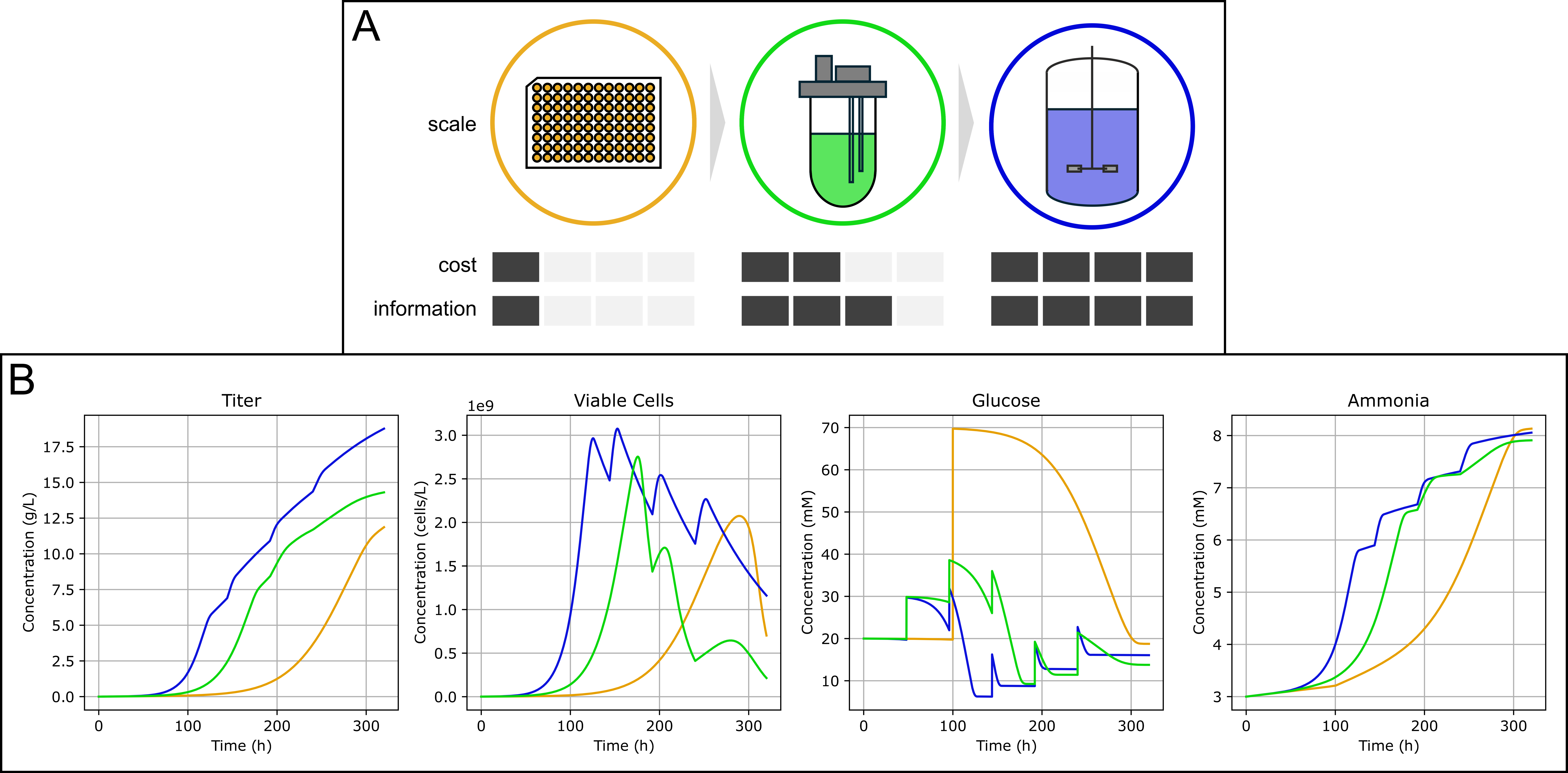
Next, we test our Bayesian optimisation framework using this in silico data. Crucially, we extend traditional optimisation by dynamically selecting both experimental scales (multi-fidelity) and batch sizes, enabling efficient exploration of the design space. In two case studies, we highlight improvements in cost efficiency and product titres, specifically compared to classical experimental designs. Our results show that integrating multi-scale data early in development leads to more robust process outcomes with fewer experiments.
Overall, this work presents a systematic screening strategy that moves beyond heuristic-driven approaches to reduce experimental burden, lower development costs, and support more informed scale-up decisions in bioprocess development.
(1) Hemmerich, J., Noack, S., Wiechert, W., & Oldiges, M. (2018). Microbioreactor systems for accelerated bioprocess development. Biotechnology journal, 13(4), 1700141.
(2) Craven, S., Shirsat, N., Whelan, J., & Glennon, B. (2013). Process model comparison and transferability across bioreactor scales and modes of operation for a mammalian cell bioprocess. Biotechnology Progress, 29(1), 186-196.
| Classification | Both methodology and application |
|---|---|
| Keywords | Bayesian optimisation, Design of Experiments, Bioprocess development |
